Diagnostic software to stratify patients on biomarkers and therapeutic targets.
Problem:
Targeted therapy has revolutionized the treatment of cancer. The efficacy of such therapy is highly dependent on target levels that may increase, decrease, or remain unchanged in the tumor compared to healthy tissue. In clinical practice, the target’s status may be tested through methods such as immunohistochemistry (IHC), which relies on a trained professional to group samples using an arbitrary cutoff. Such a semi-quantitative approach may lead to over-/under- estimation of the target expression and ineffective health interventions. Molecular assays can measure multiple biomarkers but lag in multiclass diagnostic capabilities.
Solution:
The inventor developed an original software that uses tissue biomarkers and rigorous statistical analysis to classify patients by target expression with speed and precision.
Advantages:
- Multiclass automated classification as opposed to conventional binary classification
- High diagnostic performance in single-plex and multi-plex formats
- Input from various platforms used for biomarker analysis
- The methodology can be easily applied to other biomarkers (proteins and metabolites)
- The software has diverse applications including neurology, immunology, and metabolic diseases
- The applications could be extended to animal health
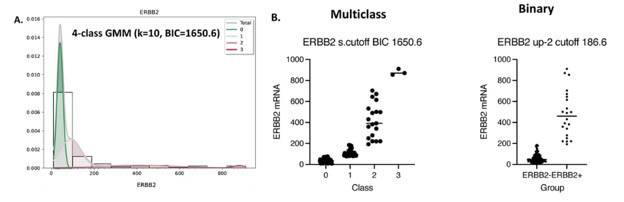
The g3mclass-assisted autoclassification on ERBB2 mRNA. (A) A total mixture model and GMM's components for the test ERBB2 mRNA were obtained with the g3mclass. The data distribution is shown as a probability density function (PDF) overlaid on a histogram and estimated by the means values, standard deviation, and weights of each component. The total GMM (gray); separate components are as follows: green—class 0 with the mean value of reference; class/es with the mean value lower than that of reference (green) and class/es with the mean value higher than that of reference (red). (B) Graphs: Examples of the multiclass (g3mclass) vs. subsequent binary classifications (conventional) of invasive breast cancer on ERBB2 mRNA.
Stage of Development:
- Software proven to run on multiple platforms (macOS, Linux, and Windows) with clinical data
Desired Partnerships:
- Non-Esclusive License
- Co-development
Case ID:
19-8759-tpNCS
Web Published:
8/12/2019
Patent Information:
| App Type |
Country |
Serial No. |
Patent No. |
File Date |
Issued Date |
Expire Date |