A method of treating Fragile X syndrome by correcting the abnormal DNA clusters causing the characteristic symptoms.
Problem:
Fragile X syndrome (FXS) affects approximately 1 in 4,000 males and 1 in 8,000 females and is characterized by symptoms such as:
- Inherited Intellectual Disability
- Anxiety
- Attention Deficit Disorder (ADD)
- Skin, Ovaries and Testes Abnormalities
FXS is caused by an excess number of regulatory CGG trinucleotides found in the Fragile X Mental Retardation 1 (FMR1) gene, termed short tandem repeats (STRs).
Compared to individuals with a normal number of STRs (30 to 40), those with over 200 STRs develop symptoms. The only current option for patients is symptom management with existing medications.
Solution:
Rather than addressing each symptom separately, this new method targets the root cause of FXS-associated symptoms by deleting the excess STRs, thus preventing abnormal DNA structures.
Technology:
The inventors linked the pathology of FXS to the formation of abnormal DNA clustering between excess STRs in the FMR1 gene and in genes of neighboring chromosomes, blocking their normal expression.
The type of DNA abnormalities and FXS symptom severity depend on STR length, with more severe disease observed in patients with over 450 repeats. Excision of the excess STRs down to 190 repeats reverses this abnormal DNA clustering and restores expression in affected genes (see Figure 1).
Shortening the STRs could be achieved using pharmacological or genetic-based methods.
Advantages:
- Reduces the number of medications FXS patients must take
- More effective than treating symptoms
- More specific than any option on the market currently
- Could be adapted to other diseases caused by STR abnormalities, such as Huntington’s disease, amyotrophic lateral sclerosis, and Friedreich’s ataxia
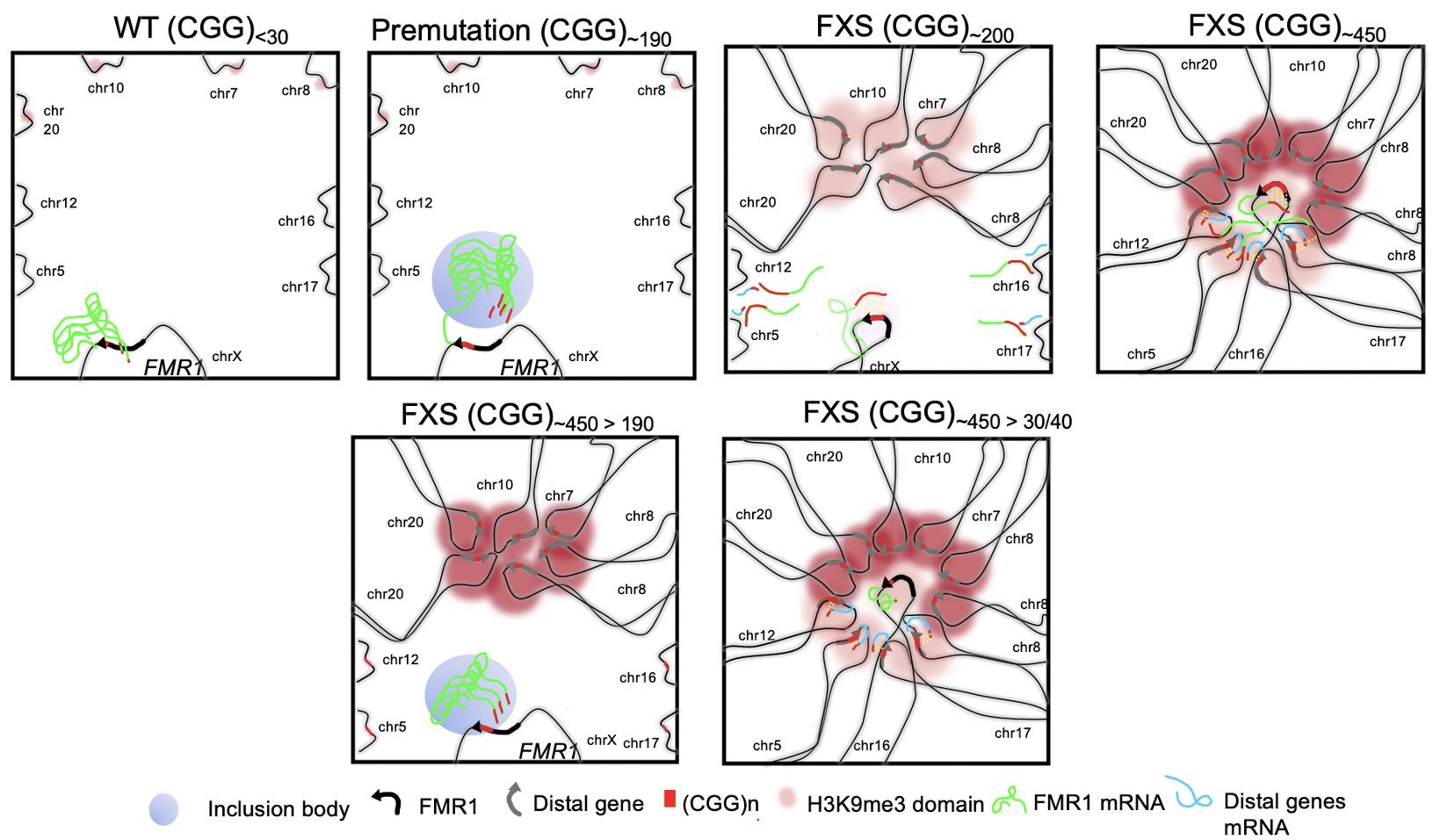
Schematic model of abnormal DNA organization among short tandem repeats (STRs) in Fragile X Syndrome. When STRs (symbolized by CGG trinucleotides, red lines in the figure) are of normal length (30-40, first box, top row), the FMR1 gene (black arrow) does not cluster with distal genes on neighboring chromosomes (gray arrows). When STRs reach 55 to 190 repeats (premutation length, second box, top row), FMR1 expression increases and forms inclusion bodies in the nucleus (blue circle) but does not interact with distal genes. When the STRs reach 200 repeats (short mutation-length, third box, top row), FMR1 expression drastically decreases, DNA instability occurs and distal genes cluster together through interactions between H3K9me3 domains (pink shading), altering their expression. When STRs reach 450 repeats (long mutation-length, fourth box, top row), the FMR1 gene forms an RNA-DNA hybrid structure (green and blue lines) which silences distal genes. Cutting the STRs from 450 to 190 repeats (first box, bottom row) reverses the abnormal interactions between FMR1 and distal genes. By contrast, cutting FMR1 STRs from 450 repeats to normal-length 30-40 repeats (second box, bottom row) does not reverse the abnormal DNA clustering.
Stage of Development:
- Target Identified
- Preclinical Discovery