Antimicrobial peptides computationally mined from the human proteome.
Problem:
Novel antibiotics are needed to counter the emergence of drug-resistant bacteria as traditional discovery approaches have not yielded new classes of antimicrobials for decades. Antimicrobial resistance (AMR) is a growing public health concern and, thus far, no significant technological advance has capitalized on this market.
Solution:
The inventors developed an algorithm to mine the human proteome for the first time as a source of antibiotics. The human proteome contains a plethora of peptides that conduct countless functions. Here, the inventors discovered that many of these peptides have antimicrobial activity outside of their primary function in the cell, and these newly discovered antimicrobial peptides (AMPs) do not readily select for bacterial resistance and display potent anti-infective activity in preclinical mouse models.
Technology:
The inventors used a computational approach to mine the human proteome for peptides with specific physicochemical properties. They intentionally screened peptides that do not resemble previously described AMPs and screened fifty-six of the discovered 2,603 candidate AMPs in vitro. Importantly, these AMPs kill bacteria by targeting the membrane and do not readily select for bacterial resistance. Most of the peptides mined displayed in vitro antimicrobial activity against eight clinically relevant pathogens ranked on the World Health Organization (WHO)’s watchlist. Of clinical importance, two candidate peptides, SCUB1-SKE25 and SCUB3-MLP2225, and a combination treatment of the two, reduced bacterial burden in an in vivo skin scarification mouse model of infection.
Advantages:
- Of 2,603 peptides mined, fifty-five were synthesized and evaluated, the majority (63.6%) of which displayed antimicrobial activity against eight clinically relevant pathogens ranked on the WHO’s watchlist
- AMPs have antimicrobial activity in vitro and in two in vivo mouse models of infection
- No significant changes in weight were observed, a proxy for toxicity
- Treatment with SCUB1-SKE25 and SCUB3-MLP2225 significantly decreased bacterial counts of P. aeruginosa and A. baumannii by three orders of magnitude respectively and combination treatment reduced bacterial counts of each pathogen by 7 and 5 orders of magnitude respectively in a mouse model of infection
- AMPs affect commensal bacteria, which can be exploited for microbiome engineering applications
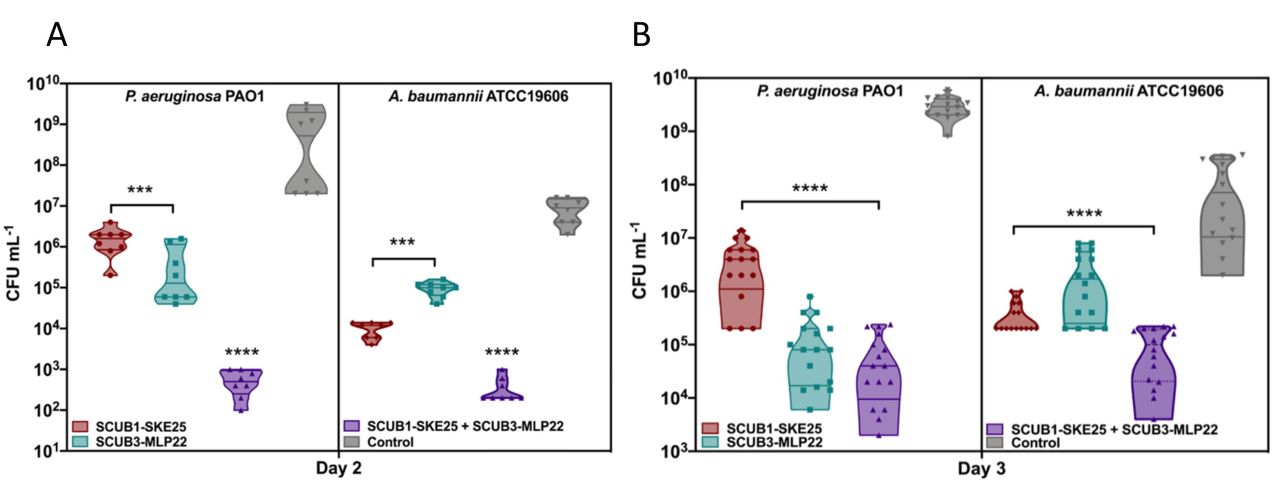
(A) Day two and (B) day three in vivo skin scarification data of AMP activity of SCUB1-SKE25 and SCUB3-MLP22 and combination treatment against P. aeruginosa PAO1 (left) and A. baumannii ATCC 19606 (right) show that these AMPs reduce bacterial burden by three orders of magnitude alone against each pathogen at days two and three and six; and, five orders of magnitude reduction on day two and seven; and, five orders of magnitude on day three respectively with combination treatment.
Stage of Development:
- Target Identified
- Pre-clinical Discovery
Reference Media:
- Torres, M.D.T. et al., Nat Biomed Eng., 2022 Jan; 6(1): 67
- Torres, M.D.T. & de la Fuente-Nunez, C., Chem Commun (Camb)., 2019 Dec 28; 55(100): 15020
- Torres, M.D.T. et al., Commun Biol., 2018 Dec 7: 1(221); 1
- University of Pennsylvania, de la Fuente Lab
Case ID:
21-9655-tpNCS
Web Published:
1/4/2024
Patent Information:
| App Type |
Country |
Serial No. |
Patent No. |
File Date |
Issued Date |
Expire Date |