A method for more targeted pre-screening for Primary Open-Angle Glaucoma (POAG) in individuals of African ancestry.
Problem:
POAG, the leading cause of irreversible blindness worldwide, currently affects 44 million individuals globally and is expected to impact 80 million people by the year 2040. Of particular concern is its disproportionate burden on individuals of African ancestry, who experience a fivefold higher prevalence than European Americans and are up to 15 times more likely to suffer from vision loss due to this condition. Despite its considerable impact, the genetic basis of POAG in African ancestry populations remains largely understudied.
POAG is often called the “silent thief of sight” because it is asymptomatic in early stages. It is estimated that approximately 50% of individuals with POAG are unaware that they have the disease. Early detection and intervention are essential because vision loss from POAG is irreversible. Treatments for POAG, though imperfect, can help to slow or halt disease progression in many patients.
There is an urgent need for improved diagnostic methods for identifying subjects of African ancestry at risk for this blinding disease. Because POAG is familial with a strong genetic component, screening by genetic testing is a viable option. Recent studies suggest that polygenic risk scores (PRS) may have clinical utility in POAG prediction and classification, but almost all PRS have been generated from variants identified in populations of European or Asian ancestry, with diminishing predictive value in African ancestry individuals.
Solution:
A genome-wide association study (GWAS) conducted by the inventors unveiled novel genetic risk factors associated with POAG, in addition to confirming known risk factors. These findings open new avenues for understanding the genetic complexities of POAG among African ancestry populations and offer potential targets for future diagnostic and therapeutic interventions. The inventors also developed a polygenic risk score for POAG in African ancestry individuals that can identify the patients at highest risk for the disease.
Technology:
The inventors conducted a GWAS where they utilized advanced genotyping and sequencing technologies to comprehensively analyze the genomes of 11,275 individuals of African ancestry. They detected 46 risk loci associated with POAG at genome-wide significance. Further functional and in silico analysis were conducted to validate the findings, identifying three likely causal variants for POAG. For individuals of African ancestry, a PRS for POAG (African ancestry individuals) outperformed a PRS from summary statistics of a much larger GWAS derived from European ancestry individuals.
Advantages:
- A total of 46 risk loci found in significant association with POAG in individuals of African ancestry.
- The large sample size allowed maximal statistical power for improved risk loci identification in African ancestry individuals.
- A significant addition to the previous POAG GWAS that predominantly contained European or Asian ancestry individuals.
- Improves targeted screening and therapeutic intervention for POAG in African ancestry individuals.
- Quantification of the genetic architecture similarities and differences between African and non-African populations for this blinding disease.
- PRS derived from our African ancestry population outperformed much larger PRS from European ancestry population when tested in African datasets, showing its clinical utility to this ancestry group.
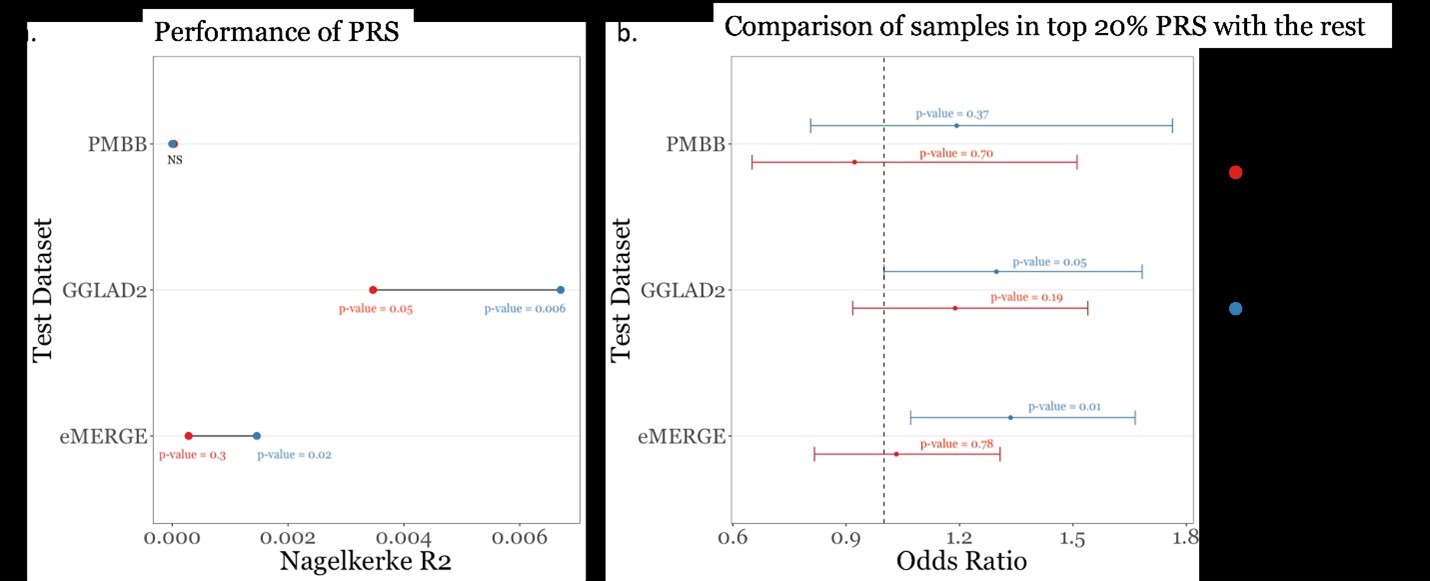
Performance of PRS generated from summary statistics from GBMI (European individuals) and the discovery mega-analysis (African individuals) in independent datasets of African ancestry individuals. Summary statistics from GBMI (European individuals) and discovery mega-analysis (African individuals) were each used to create a PRS. Each PRS was measured in three independent datasets of African ancestry individuals (eMERGE, GGLAD-2, PMBB). Fig. 6A: The Nagelkerke R2 is shown on the x-axis and the test datasets are shown on the y-axis, with the P-value corresponding to the Nagelkerke p-value. Fig. 6B: The odds ratio (x-axis) was calculated by comparing individuals in top 20% PRS with rest of the test datasets (y-axis). The P-value corresponds to regression p-value.
Case ID:
22-10097-TpNCS
Web Published:
10/16/2024
Patent Information:
| App Type |
Country |
Serial No. |
Patent No. |
File Date |
Issued Date |
Expire Date |